
Key Parameters and Techniques for Faster Processing
Uniform Manifold Approximation and Projection (UMAP) is a powerful algorithm widely used for dimensionality reduction and visualizing complex data. Its ability to retain data structure makes it an excellent tool for high-dimensional datasets.
However, tuning UMAP for optimal performance can be challenging, especially with large datasets where processing speed and memory are crucial. In this article, we’ll walk through the key parameters and techniques that will help you make UMAP faster and more efficient.
Understanding UMAP’s Core Parameters for Speed
Adjusting n_neighbors to Control Locality
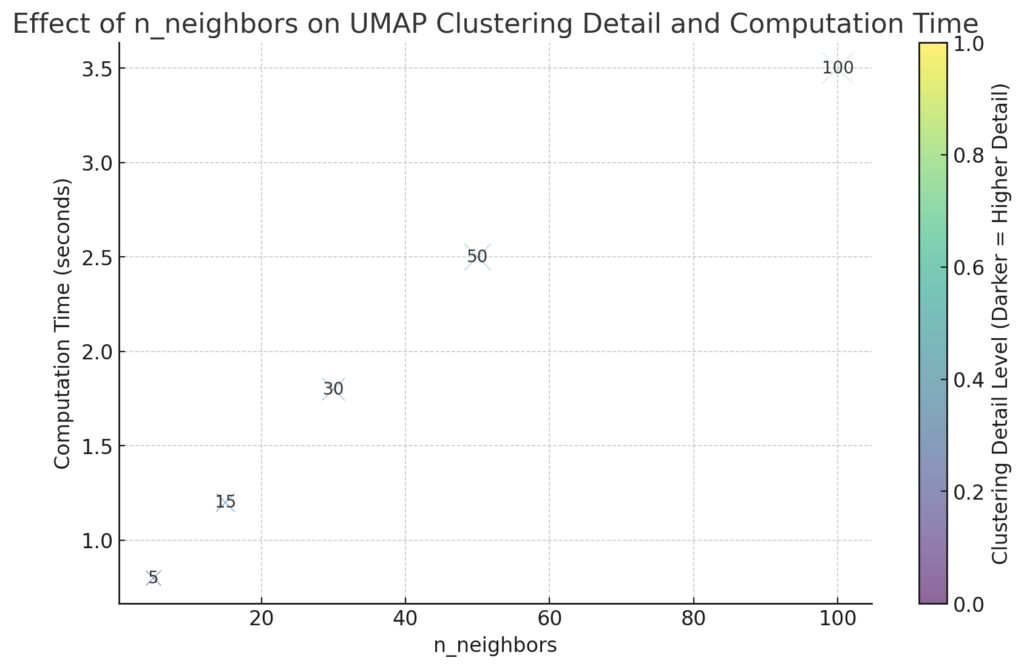
The n_neighbors parameter in UMAP defines the number of neighboring points considered when calculating the manifold structure. It influences both the quality and speed of UMAP projections.
- Lower values of
n_neighborscapture finer details and can speed up computation but may lose the global structure. - Higher values provide more stability but increase computational demands.
For performance tuning, start with n_neighbors between 5 and 30, depending on the dataset’s size and complexity. Reducing it in initial runs can help you find an ideal balance between performance and structure retention.
Setting min_dist to Control Point Clustering
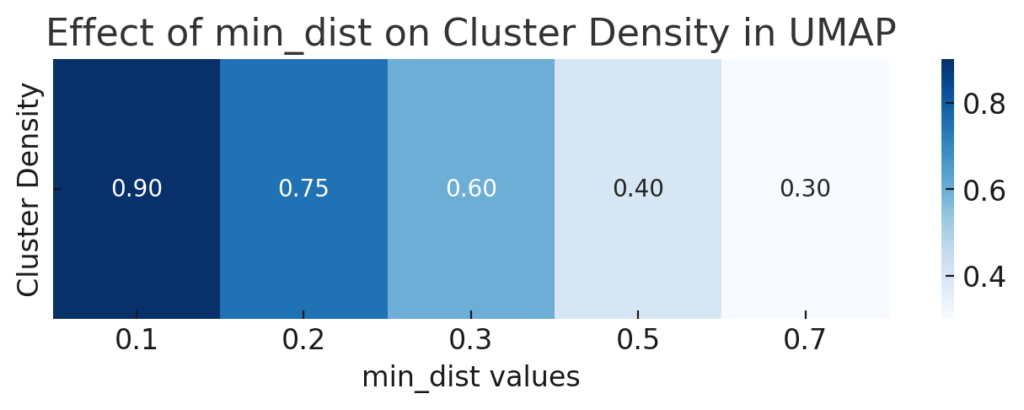
UMAP’s min_dist parameter affects how closely points in the projection are allowed to be. While it doesn’t directly impact processing speed, it influences how data clusters, which can indirectly affect performance by modifying how much detail needs to be retained.
- Lower values create tighter clusters, often revealing dense, intricate structures.
- Higher values create a more spread-out, smoother layout, which can simplify processing.
For faster, less detailed processing, try starting with a min_dist of 0.3 or higher. Lower values can be used later if you need more granular data structures.
Tweaking metric for Similarity Calculations
The distance metric used to compute similarity between points can impact UMAP’s speed significantly. Euclidean distance is the default, but other metrics, like cosine or Manhattan, might better fit certain types of data. However, Euclidean distance is computationally efficient and well-suited to most datasets.
For speed optimization, stick with Euclidean unless you have a strong reason to switch. If your data has inherent directional or sparse properties, experimenting with metrics like cosine or correlation may yield better performance while retaining quality.
Optimizing Computational Techniques
Using Smaller Subsets for Initial Tests
When working with UMAP, start with a small subset of your data for initial parameter tuning. By sampling 10-20% of your data, you can quickly assess which parameters lead to faster results without waiting for the entire dataset to process.
This technique is particularly helpful in:
- Determining approximate values for
n_neighborsandmin_dist - Testing different metrics and configurations
- Reducing the trial-and-error time in fine-tuning
Once you find effective parameters, apply them to the full dataset for more efficient and accurate results.
Adjusting n_components for Lower Dimensions
The n_components parameter defines the number of dimensions in the UMAP output. While a higher number of dimensions may be needed for some analysis types, setting n_components to 2 or 3 for visualization purposes will usually be sufficient and can significantly reduce computation time.
For exploratory data analysis or visualizations, stick with 2 or 3 components to maximize performance. Only increase n_components if your analysis specifically requires it, as each added dimension increases processing demands.
Leveraging Hardware and Parallelization
Utilizing GPU Acceleration
For substantial datasets, UMAP can be significantly accelerated by GPU processing. Using UMAP’s GPU-compatible implementations, like UMAP with RAPIDS, can drastically reduce computation time on supported hardware.
To make use of GPU acceleration:
- Install the RAPIDS library and compatible UMAP package.
- Enable GPU processing by selecting the right library version or setting the GPU flag if using a library that supports it.
Using GPU can speed up UMAP by 10x or more, particularly on large datasets with high-dimensional features.
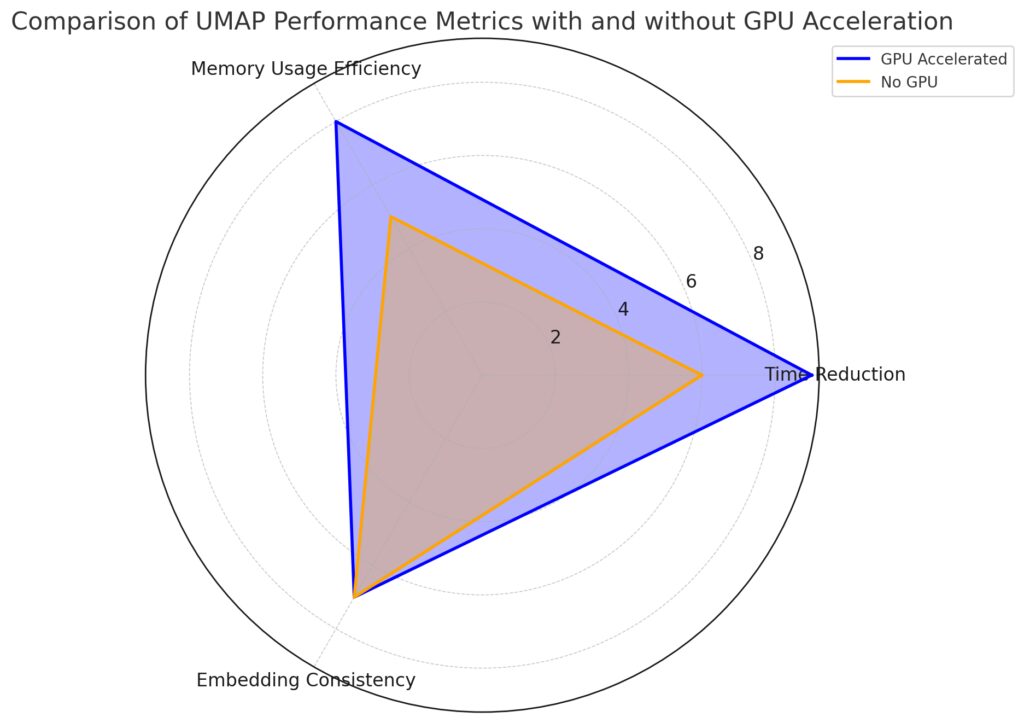
Memory Usage Efficiency: GPU optimization enhances memory usage, indicated by a higher score with GPU.
Embedding Consistency: Both configurations achieve similar embedding quality, maintaining consistency.
Parallelizing Computations with n_jobs
The n_jobs parameter in UMAP controls the number of CPU cores used during processing. By setting n_jobs to -1, you allow UMAP to utilize all available cores, which can significantly speed up processing on multicore systems.
For optimal performance:
- Set
n_jobsto-1to enable full parallelization. - Monitor your system resources to ensure that parallel processing doesn’t overload memory, especially on machines with limited RAM.
If you’re working on a server or workstation with multiple cores, parallelizing can reduce computation time by distributing the workload effectively.
Optimizing UMAP with Data Preprocessing Techniques
Dimensionality Reduction Before UMAP
Reducing the number of dimensions in your data before applying UMAP can significantly improve processing time without sacrificing much in terms of final projection quality. Consider using PCA (Principal Component Analysis) as a preliminary step.
Steps for preprocessing with PCA:
- Apply PCA to reduce dimensions to a manageable level (e.g., from thousands of dimensions to 50-100).
- Use this reduced data as input for UMAP.
By applying PCA first, you can drastically cut down the number of computations that UMAP needs, resulting in faster runtime without a significant loss in detail.
Data Scaling for Consistent Distance Calculations
Scaling your data can also improve UMAP’s performance and accuracy. Inconsistent data scales may cause UMAP to overemphasize certain features, which can slow down calculations and reduce the quality of projections.
- Use standardization or normalization to ensure all features have a similar scale.
- For datasets with varied feature types, consider scaling by feature importance to emphasize the most critical features.
Proper scaling can help UMAP create a more accurate manifold, thus optimizing performance and producing clearer, more consistent results.
Incorporating these key parameter adjustments and processing techniques will help you maximize UMAP’s efficiency, especially when handling complex datasets. By understanding and tuning the core parameters like n_neighbors, min_dist, and n_components, you can strike the perfect balance between processing speed and data structure quality. With the right setup, UMAP becomes not just a tool for dimensionality reduction, but a streamlined part of your data analysis pipeline.
Fine-Tuning UMAP for Large Datasets
Applying Sparse Matrices for High-Dimensional Data
When working with high-dimensional data, especially those with sparse elements (such as text data in NLP or certain types of genomic data), leveraging sparse matrix representations can speed up UMAP significantly. Sparse matrices store only non-zero elements, reducing memory usage and processing time.
To optimize UMAP with sparse matrices:
- Convert your data to a sparse matrix format if it’s not already.
- Ensure that UMAP parameters like
metricandtransformsettings are compatible with sparse matrices, as some configurations might require dense data formats.
Using sparse matrices can lead to faster computations without needing to downsample your data, making it an excellent option for data types with a lot of zero or empty values.
Reducing Iterations with n_epochs
UMAP’s n_epochs parameter controls the number of optimization iterations. While higher values may produce more refined results, lowering n_epochs can significantly reduce computation time.
For faster processing:
- Start with a low
n_epochsvalue, around 100 to 200. This can provide a quick but rough projection, useful for initial analysis. - If needed, gradually increase
n_epochsto see how much further refinement is possible without significantly impacting performance.
Using a reduced n_epochs in early stages allows you to analyze UMAP projections faster. Once you’re satisfied with the general layout, you can decide whether higher epochs are worth the extra processing time.
Tuning learning_rate for Efficient Convergence
The learning_rate parameter determines how fast UMAP converges toward a solution. Higher learning rates can speed up computation by moving faster toward optimal solutions, but may cause instability or poor quality in the final projection.
- Start with the default learning rate, around 1.0.
- If UMAP runs slowly or converges inefficiently, try increasing
learning_rateup to 2 or 5. This often helps find a quicker solution, especially on large datasets.
Higher learning_rate values can save time in many cases, but be cautious; a very high value may degrade the quality of your results, so it’s best to experiment in small increments.
Downsampling for a Quick Overview
If you’re working with an exceptionally large dataset, consider downsampling as a preliminary step. Running UMAP on a representative subset of your data can quickly provide insights into parameter settings, likely clusters, and data structure.
To use downsampling effectively:
- Select a random or stratified sample of 10-25% of your dataset.
- Run UMAP on the sampled data with standard parameters and note the results.
- Apply the learned parameters or adjust them when running UMAP on the full dataset.
Downsampling allows you to test hypotheses and identify useful configurations quickly. It also reduces the strain on memory and CPU, making the entire process more manageable.
Advanced Techniques for Improved UMAP Speed
Enabling Approximation for Faster Calculations
For high-dimensional data, approximation techniques can drastically reduce computation time. UMAP offers several built-in options for approximation, which use shortcuts or sampling to arrive at results more quickly.
- Enable approximate nearest neighbor algorithms by setting the
metricparameter to options like ‘approximate’ if available. UMAP will automatically choose the most efficient approximation for your data. - Use annoy or pynndescent, two popular libraries for approximate nearest neighbor calculations compatible with UMAP.
Approximation methods may yield slightly less accurate projections but provide a significant boost in performance. This trade-off can be valuable when your focus is exploratory analysis rather than final output.
Utilizing Batch Processing for Sequential Analysis
For extremely large datasets, batch processing allows you to process data in smaller chunks sequentially. This reduces memory demands, especially useful when working with limited resources or very high-dimensional data.
To enable batch processing:
- Divide your data into manageable batches (10-20% of the data).
- Run UMAP on each batch independently, saving intermediate results for comparison.
- Use embedding stitching techniques if you need to combine batch results for a final projection.
Batch processing is useful when GPU or RAM resources are limited. Although stitching together embeddings from batches can be complex, this technique makes it possible to scale UMAP across larger datasets.
Monitoring UMAP Performance Metrics
Tracking Computation Time and Memory Usage
UMAP’s performance can vary depending on data type, size, and hardware, so tracking runtime and memory usage is essential. Regular monitoring helps you catch bottlenecks and adjust parameters for a more efficient run.
Use profiling tools like:
- Python’s
timeitmodule to track computation time. - Memory profiling tools like
memory_profilerto monitor memory usage during runs.
These tools help identify where UMAP slows down and enable you to make data-driven adjustments to improve processing speed.
Analyzing UMAP Quality with Silhouette and Trustworthiness Scores
Besides speed, it’s crucial to assess UMAP’s output quality. Silhouette scores and trustworthiness scores are popular metrics for validating the clustering and data structure retained in the projection. Low values may indicate the need for parameter tuning or adjustments to UMAP’s configuration.
- Use Silhouette scores to assess cluster quality—higher scores reflect well-defined clusters.
- Trustworthiness scores measure how well UMAP preserves the original data relationships.
By evaluating these metrics alongside runtime, you can better understand the balance between quality and speed in your UMAP configuration.
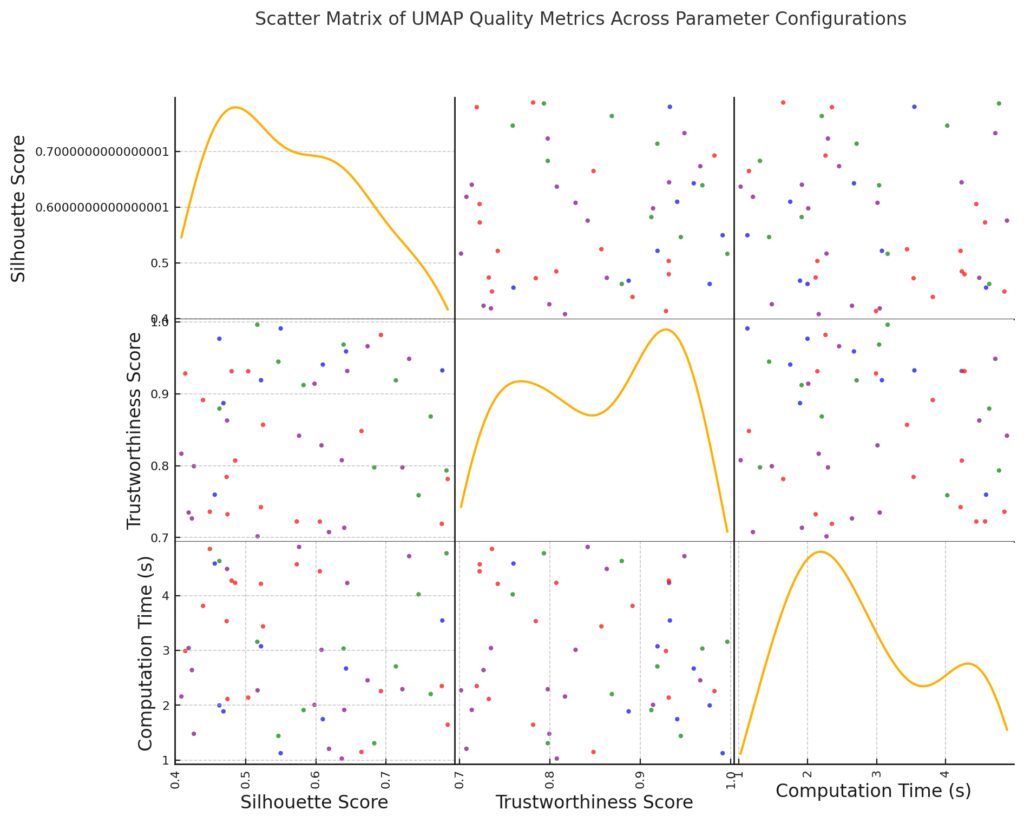
Silhouette Score and Trustworthiness Score: Indicate clustering quality and the preservation of local structure, respectively.
Computation Time (s): Reflects the computational efficiency for each configuration.
Color-Coding: Each configuration is uniquely color-coded (blue, green, red, and purple) to help differentiate the parameter settings.
This matrix allows for visual comparisons of how different configurations affect the balance between clustering quality and computation time, making it easier to identify efficient configurations that maintain high-quality embeddings
With these advanced techniques and monitoring strategies, you can optimize UMAP for faster and more efficient processing on both small and large datasets. Tuning parameters like n_epochs, learning_rate, and leveraging techniques like approximation and batch processing are essential for scaling UMAP to handle complex, high-dimensional data efficiently.
Leveraging UMAP Transformations for Scalable Analysis
Using the transform() Function for New Data
UMAP offers a transform() function to apply learned embeddings to new data, enabling you to update projections without reprocessing the entire dataset. This is particularly useful for applications requiring frequent updates, such as time-series data or real-time analysis.
To use transform() effectively:
- Run UMAP on the initial dataset and save the model.
- Use
umap_model.transform(new_data)on incoming data to integrate it with the original embedding.
The transform() function saves time by allowing you to extend UMAP’s projections as new data arrives, making it ideal for scalable applications like streaming data or incremental updates.
Optimizing Incremental Training for Continuous Data Streams
If your data is dynamic and constantly evolving, incremental training provides a way to keep embeddings updated without re-running UMAP from scratch. While UMAP is not natively designed for incremental updates, certain implementations and libraries, such as UMAP-learn with supervised settings, allow for incremental adjustments.
For incremental training:
- Set up a UMAP pipeline with batch updates on small, new data chunks.
- Fine-tune parameters like
learning_rateandmin_distto control how quickly UMAP integrates new data into the existing embedding space.
Incremental training lets you maintain up-to-date embeddings, making UMAP adaptable to applications like anomaly detection or evolving trend analysis.
Final Thoughts on Optimizing UMAP for High Performance
Optimizing UMAP for speed and efficiency requires a balance between parameter tuning, data preprocessing, and computational techniques. Key parameters like n_neighbors, min_dist, and learning_rate offer initial control over processing time and projection quality, while advanced techniques—like approximation, batch processing, and sparse matrices—allow UMAP to handle larger datasets with less computational strain.
Incorporating UMAP with GPU acceleration, parallel processing, or incremental learning can further extend its capability, especially for applications where data is vast, high-dimensional, or frequently updated. By employing these methods, you’ll achieve faster, more accurate, and adaptable UMAP embeddings suitable for various data science and machine learning applications.
Ultimately, understanding the trade-offs between speed and quality, and monitoring performance metrics, will help you fine-tune UMAP for real-world, high-performance applications in data analysis, visualization, and beyond.
FAQs
What’s the benefit of preprocessing data with PCA before UMAP?
Applying PCA (Principal Component Analysis) before UMAP reduces dimensionality, often resulting in faster UMAP computations. Reducing thousands of dimensions to 50-100 using PCA maintains most of the variance while making UMAP’s job easier and faster on complex datasets.
How does batch processing improve UMAP performance?
Batch processing allows you to process large datasets in smaller chunks, reducing memory usage and CPU demand. This technique is useful if your system has limited resources or if your dataset is too large to process in a single run. By processing one batch at a time, you avoid memory overload, and for applications that don’t require the full dataset at once, it offers faster incremental insights.
Can I use different metrics in UMAP to improve performance?
Yes, UMAP supports various distance metrics to calculate similarity between points. The default, Euclidean distance, is computationally efficient and works well with most datasets. However, for text data or high-dimensional sparse data, metrics like cosine or correlation may be more appropriate, though they may increase computation time slightly. Testing different metrics based on your data type can improve both the quality and speed of UMAP embeddings.
Is there a trade-off between n_neighbors and min_dist in UMAP?
Yes, n_neighbors and min_dist work together to shape UMAP’s projection. Generally, lower n_neighbors and higher min_dist values create a broader, less detailed representation, which can process faster. In contrast, higher n_neighbors and lower min_dist will capture more structure and detail, increasing processing demands. Experimenting with both parameters will help you find the right balance between capturing data detail and improving speed.
Can I use UMAP for supervised tasks, and does it impact performance?
Yes, UMAP can be used in a supervised mode by passing class labels to guide the embedding. This can improve projection quality by ensuring that points within the same class are grouped closely. However, supervised UMAP can slightly increase processing time, especially with large labeled datasets, since the algorithm has to consider additional class structure.
How does approximation impact the quality of UMAP projections?
UMAP can use approximate nearest neighbor algorithms to speed up calculations, which can improve processing time substantially with minimal effect on quality. While approximation might lead to slight variations in the embedding, the impact is usually minimal for exploratory data analysis. However, for highly precise applications, you may want to run UMAP without approximation, albeit at the cost of slower computation.
What tools can I use to monitor UMAP’s memory and runtime performance?
Tools like timeit in Python can measure runtime, while memory_profiler can track memory usage during UMAP runs. These tools allow you to pinpoint bottlenecks and high memory usage in your UMAP configuration, helping you optimize settings or consider techniques like downsampling or sparse matrices to improve performance.
How do sparse matrices work with high-dimensional data in UMAP?
Sparse matrices only store non-zero elements, making them highly efficient for high-dimensional data with many zeros, such as word embeddings in NLP. Using sparse matrices with UMAP reduces memory requirements and often improves computation speed, as UMAP processes fewer non-zero elements compared to dense matrices. Converting your data to a sparse format before running UMAP can be a practical step for very large or sparse datasets.
What are the advantages of using UMAP with incremental training?
While UMAP doesn’t support true incremental learning natively, certain UMAP extensions allow incremental updates by processing small batches of new data. This approach is beneficial in real-time data scenarios where new data points frequently arrive, such as time-series or streaming applications. Incremental training allows UMAP to adapt without reprocessing the entire dataset, making it a viable solution for continuous analysis applications.
When should I consider downsampling my dataset for UMAP?
Downsampling is recommended when your dataset is exceptionally large, and you need a quick look at general structure before fine-tuning. By working with a representative subset, you can adjust key parameters without waiting for the entire dataset to process, thus speeding up the tuning process. Downsampling is especially effective for large, high-dimensional datasets, as it reduces memory load and computational requirements while still giving a reliable indication of clustering patterns.
What is the effect of using a high learning_rate in UMAP?
A higher learning_rate allows UMAP to converge faster by making larger adjustments with each iteration. This can reduce computation time, especially for larger datasets, but it may also lead to less stable embeddings if set too high. For optimal performance, start with the default (1.0) and increase gradually to 2 or 5 to see if it improves speed without sacrificing quality. Avoid setting it too high, as this can create distortions in the data projection.
Can UMAP handle real-time data streams effectively?
UMAP can be adapted to real-time or streaming data by leveraging the transform() function for new data points. While UMAP doesn’t natively support streaming, you can preprocess initial embeddings and apply the transform() method to incoming data, integrating it into the original embedding space without re-running the full algorithm. This is ideal for applications where the dataset is dynamic, such as real-time clustering or continuous anomaly detection.
How does PCA help with preprocessing before UMAP?
Applying PCA (Principal Component Analysis) as a preprocessing step before UMAP reduces the data’s dimensionality, which can drastically lower computational demands. PCA captures the main variance in the data, making it possible to reduce thousands of dimensions to a more manageable number (e.g., 50–100) before running UMAP. This results in faster UMAP processing while still retaining essential data structure and relationships.
What are the memory benefits of using sparse matrices in UMAP?
Sparse matrices store only the non-zero values in a dataset, making them extremely efficient for sparse high-dimensional data (like text data with one-hot encodings). By using sparse matrices, UMAP saves memory and processes fewer elements, speeding up computation. This format is especially beneficial for handling large, sparse datasets, as it reduces both memory requirements and computation time without losing data structure.
What is the difference between n_epochs and n_neighbors in UMAP?
n_epochs controls the number of training iterations and influences how finely UMAP optimizes the embedding. Lower n_epochs values (100–200) yield faster but less refined embeddings, suitable for quick visualizations. n_neighbors, on the other hand, controls the size of the local neighborhood UMAP considers around each point, influencing the level of detail captured in the data. Higher n_neighbors values provide more stability in structure but may slow down processing.
How does the metric parameter affect UMAP performance and quality?
The metric parameter defines the distance measure UMAP uses to determine similarity between data points. Euclidean is the default and fastest for most purposes, but for specialized data, like text or genetic data, metrics like cosine or correlation may provide better embeddings. However, non-Euclidean metrics tend to increase computation time, so balance between optimal distance metric and runtime when selecting a non-default metric.
Is it possible to parallelize UMAP for faster processing?
Yes, UMAP can use multiple CPU cores with the n_jobs parameter, which can speed up processing significantly, particularly for large datasets. Setting n_jobs=-1 enables UMAP to utilize all available cores. This allows you to distribute the workload and can improve runtime without changing the algorithm itself. If running on a multicore system, this parallelization can provide a noticeable boost to performance.
What is the purpose of the min_dist parameter, and when should I adjust it?
The min_dist parameter controls how close points can be in the UMAP embedding. It doesn’t directly affect speed, but it does impact the spread and tightness of clusters in the final visualization. Lower values (e.g., 0.1) create tight clusters, emphasizing local relationships, while higher values (e.g., 0.3 or above) result in a more spread-out embedding that captures broader data structure. For an overview of general patterns in data, start with a higher min_dist. Adjust it lower if you need to visualize fine-grained, closely packed clusters.
Can UMAP handle categorical data, and does this affect performance?
UMAP can process categorical data by encoding it as one-hot or binary vectors. However, this approach often increases dimensionality significantly, which can slow down processing. To optimize performance, try encoding categories as low-dimensional embeddings instead, or use targeted feature engineering to reduce dimensionality before running UMAP. High-dimensional categorical data might also benefit from using sparse matrices to further improve efficiency.
How does UMAP compare to t-SNE in terms of performance?
UMAP is generally faster and more scalable than t-SNE (t-Distributed Stochastic Neighbor Embedding), especially on large datasets. UMAP has a better balance of preserving both local and global structure, which makes it suitable for high-dimensional data while maintaining speed. While t-SNE can produce high-quality embeddings, it is typically slower and less adaptable to large or streaming data compared to UMAP, especially without parallel processing or GPU support.
What are the best practices for using UMAP with imbalanced datasets?
For imbalanced datasets, carefully tuning n_neighbors and min_dist can help maintain meaningful clusters even when certain classes are underrepresented. Lowering n_neighbors allows smaller clusters to form, which can make minority classes more visible in the embedding. You might also consider oversampling techniques or applying supervised UMAP (by passing labels) to enhance the clustering of less frequent classes, though these steps may increase computation time.
Can I use UMAP with both numerical and categorical data?
Yes, UMAP can handle mixed data types by preprocessing categorical variables separately and merging them back with numerical data. For instance, you might apply one-hot encoding to categorical features and scaling to numerical features. Keep in mind that this may increase dimensionality, so using feature selection or PCA can be useful before running UMAP to prevent slowdowns and memory overload.
Does transform() work with new data that has different distributions?
UMAP’s transform() function is designed to apply learned embeddings to new data points, which works best when the new data’s distribution is similar to the original dataset. If new data has a very different distribution, results may be less reliable, as UMAP doesn’t retrain on the new data. For substantial distribution shifts, consider re-running UMAP on a combined dataset that includes both old and new data to maintain embedding quality.
How does using a higher n_components affect UMAP’s processing speed?
n_components specifies the dimensionality of the output. While UMAP is typically used for 2D or 3D embeddings, higher dimensions can be used for complex analysis. However, each additional dimension increases computation and memory requirements. For exploratory data analysis, 2 or 3 components is usually sufficient and optimal for speed. Higher dimensions should be reserved for specific tasks where detailed data representation is critical, despite the additional computational load.
Are there any specific applications where UMAP performs particularly well?
UMAP is highly effective in applications requiring dimensionality reduction for visualizations, such as genomics, text embeddings, image data, and anomaly detection. Its balance of speed and structure preservation makes it a go-to for exploratory analysis, especially in large and high-dimensional datasets. It also performs well in real-time and streaming data applications, where you can use transform() for continuous embedding updates. For classification tasks, supervised UMAP can create embeddings that help separate classes effectively, aiding in visualizations for clustering and segmentation.
Resources
Official UMAP Documentation
The official UMAP documentation provides comprehensive information on the algorithm’s parameters, usage, and technical details. It’s a great starting point for understanding core parameters like n_neighbors, min_dist, and n_epochs and how they influence performance.
RAPIDS and UMAP for GPU Acceleration
For large datasets, GPU acceleration with RAPIDS is essential. The RAPIDS UMAP library offers a GPU-compatible implementation of UMAP, which drastically speeds up processing on supported hardware. The documentation explains setup, parameter options, and performance benchmarks for UMAP on GPU.
UMAP with Scikit-Learn Integration
UMAP integrates well with Scikit-Learn, providing an accessible framework for UMAP alongside other dimensionality reduction techniques. This integration is helpful for users familiar with Scikit-Learn, allowing easy comparisons of UMAP with PCA, t-SNE, and other methods.
Research Paper on UMAP
The original UMAP research paper by Leland McInnes, John Healy, and James Melville is an essential resource for understanding the mathematical foundations and motivations behind UMAP. It provides an in-depth explanation of UMAP’s manifold learning, neighborhood construction, and optimization steps.
PyData and UMAP Video Tutorials
The PyData YouTube channel hosts presentations and tutorials on UMAP, often featuring experts like Leland McInnes, one of UMAP’s authors. These videos offer practical insights into UMAP parameter tuning, use cases, and performance optimization techniques.
GitHub Repository for UMAP
The UMAP GitHub repository contains the source code, issues, and discussions on UMAP development. Reviewing the code and issues can provide a deeper understanding of customizing UMAP for specific performance needs, implementing new metrics, and keeping track of updates and optimizations.
Memory and Performance Profiling Tools
For tracking UMAP’s performance, tools like Memory Profiler and timeit in Python are essential. Memory Profiler allows you to monitor memory usage, and timeit can help you measure UMAP’s processing time on various settings, which is valuable for performance tuning.
Tutorials and Blogs on UMAP Parameter Tuning
There are several high-quality blog tutorials on tuning UMAP for specific datasets and applications. Some standout resources include:
- Towards Data Science: Numerous UMAP articles with step-by-step guides on parameter selection for tasks like text embeddings and image clustering.
- Distill.pub: Visual explanations and articles on dimensionality reduction techniques, including UMAP and t-SNE, to understand when to use each technique.
Papers and Studies Comparing UMAP with Other Algorithms
Research comparing UMAP to other dimensionality reduction methods like t-SNE, PCA, and autoencoders provides useful insights into performance trade-offs and use cases. This comparison study on UMAP and t-SNE can help you understand where UMAP has advantages in speed and structure preservation.